Areas of research
The National Center for Gene Research (NCGR) of Chinese Academy of Sciences (CAS) is now focused on rice genome re-sequencing for genotyping, genome-wide association studies (GWAS) to identify common genetic factors, comparative genome and transcriptome analysis of indica and japonica, and rice functional genomics as well. Our research is divided into four main areas: rice genetics, bamboo genetics, Miscanthus genetics and bioinformatics.
- Rice genetics
- Bamboo genetics
- Miscanthus genetics
- Bioinformatics
Background: Rice is one of the most important crops and an excellent monocotyledonous model plant. Since 2003, with the completion of rice genome sequences and genetic and genomic resources, rice has become a model for comparative genomics, genome annotations and functional genomics. The essential steps toward deciphering the sequenced genome are to develop complete and accurate maps of actively transcribed regions during rice development, and to generate more and more genome-wide rice resources and datasets (including mutants, genetic markers, small RNAs and so on). Recently, the development of the next-generation high-throughput DNA sequencing technologies provided a novel method for these different researches.
Researches:
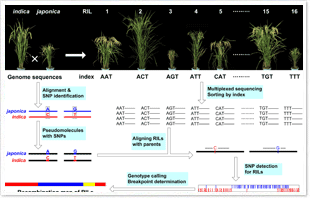
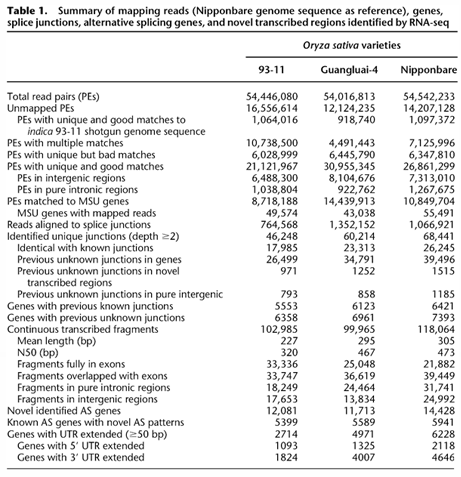 1.The next-generation sequencing technology coupled with the growing number of genome sequences opens the opportunity to redesign genotyping strategies for more effective genetic mapping and genome analysis. We have developed a high-throughput method for genotyping recombinant populations utilizing whole-genome resequencing data generated by the Illumina Genome Analyzer.
1.The next-generation sequencing technology coupled with the growing number of genome sequences opens the opportunity to redesign genotyping strategies for more effective genetic mapping and genome analysis. We have developed a high-throughput method for genotyping recombinant populations utilizing whole-genome resequencing data generated by the Illumina Genome Analyzer.
2.The RNA-seq approach was applied to analyze the global transcriptome of rice at the best possible resolution. We globally investigate alternative splicing (AS) patterns, novel transcriptional active regions (nTARs), differentially expressed genes (DEGs), and coding region comparisons between subspecies to improve the annotation of the rice genome.
Selected Publications:
China National Rice Research Institute
National Key Laboratory of Crop Genetic Improvement
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Researches:


2.The RNA-seq approach was applied to analyze the global transcriptome of rice at the best possible resolution. We globally investigate alternative splicing (AS) patterns, novel transcriptional active regions (nTARs), differentially expressed genes (DEGs), and coding region comparisons between subspecies to improve the annotation of the rice genome.
High-throughput genotyping by whole-genome resequencing.
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B.
Genome Res. 2009; 19: 1068-76.
PMID: 19420380
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B.
Genome Res. 2009; 19: 1068-76.
PMID: 19420380

Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq.
Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Li W, Huang X, Han B.
Genome Res. 2009; 20: 1238-49.
PMID: 20627892
Collaborations: Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Li W, Huang X, Han B.
Genome Res. 2009; 20: 1238-49.
PMID: 20627892

China National Rice Research Institute

National Key Laboratory of Crop Genetic Improvement

Institute of Genetics and Developmental Biology, Chinese Academy of Sciences

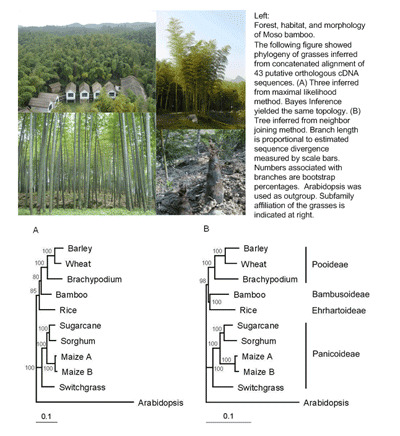 Background: We rely on grasses more than any other groups of plants for food and potential renewable energy. With the number of genome sequences growing much more rapidly than any other plant family, the grass family (Poaceae) becomes an ideal system for comparative studies of gene and genome structure and function. To develop the system, it is critical to accumulate genomic resources for all major lineages of the grass family. The majority of large subfamilies of grasses have already had a great deal of genomic or expressional data available primarily because they possess crop species. The only exception is the subfamily of bamboos, Bambusoideae, which contains more than 1,000 species but has little data available in DNA or protein sequences databases. This creates a major missing link in the grass family for comparative genomics.
Background: We rely on grasses more than any other groups of plants for food and potential renewable energy. With the number of genome sequences growing much more rapidly than any other plant family, the grass family (Poaceae) becomes an ideal system for comparative studies of gene and genome structure and function. To develop the system, it is critical to accumulate genomic resources for all major lineages of the grass family. The majority of large subfamilies of grasses have already had a great deal of genomic or expressional data available primarily because they possess crop species. The only exception is the subfamily of bamboos, Bambusoideae, which contains more than 1,000 species but has little data available in DNA or protein sequences databases. This creates a major missing link in the grass family for comparative genomics.Researches:
As the first step of the bamboo genome sequence projects, we cloned and sequenced more than ten thousand putatively unique FLcDNA derived primarily from vegetative tissues of Moso bamboo, Phyllostachys heterocycla cv. Pubescens, a large woody bamboo in Asia.
Selected Publications:
Genome-wide characterization of the biggest grass, bamboo, based on 10,608 putative full-length cDNA sequences.
Peng Z, Lu T, Li L, Liu X, Gao Z, Hu T, Yang X, Feng Q, Guan J, Weng Q, Fan D, Zhu C, Lu Y, Han B, Jiang Z.
BMC Plant Biol. 2010; 10: 116.
PMID: 20565830
Collaborations: Peng Z, Lu T, Li L, Liu X, Gao Z, Hu T, Yang X, Feng Q, Guan J, Weng Q, Fan D, Zhu C, Lu Y, Han B, Jiang Z.
BMC Plant Biol. 2010; 10: 116.
PMID: 20565830

International Network for Bamboo and Rattan

Chinese Academy of Forestry

Background: Miscanthus is one of a number of renewable energy crops that could be used as a fuel for power stations or heating systems. Miscanthus is the sterile, hybrid forms that are of value agriculturally. It is a woody, perennial grass originating from Asia that grows fast and after the second year of establishment, provides an annual harvest. China possesses the biggest biomass production and owns abundant genetic diversity and plant ecology of Miscanthus.
Researches:
Relying on the large resources and by using the high-throughput sequencing technology and molecular breeding approaches, we plan to widely investigate, collect and evaluate the status of genetic diversity and ecology of Miscanthus in China.
Collaborations:
Institute of Botany, CAS
Wuhan Botanical Garden, CAS
Researches:

Relying on the large resources and by using the high-throughput sequencing technology and molecular breeding approaches, we plan to widely investigate, collect and evaluate the status of genetic diversity and ecology of Miscanthus in China.
Collaborations:
Institute of Botany, CAS

Wuhan Botanical Garden, CAS

The Bioinformatics programme develops and applies methods to process, store and analyze data generated by high-throughput projects.
Researches:

1.The Oryza sativa L. indica subspecies is the most widely cultivated rice. During the last few years, we have collected over 20,000 putative full-length cDNAs and over 40,000 ESTs isolated from various cDNA libraries of two indica varieties Guangluai 4 and Minghui 63. A database of the rice indica cDNAs was therefore built to provide a comprehensive web data source for searching and retrieving the indica cDNA clones.
Related website
2.With the availability of rice and sorghum genome sequences and ongoing efforts to sequence genomes of other cereal and energy crops, the grass family (Poaceae) has become a model system for comparative genomics and for better understanding gene and genome evolution that underlies phenotypic and ecological divergence of plants. We developed a Moso bamboo genomic resource database that so far contained the sequences of 10,608 putative FL-cDNAs and nearly 38,000 expressed sequence tags (ESTs) generated in this study.
Related website
3.We applied the RNA-seq approach to analyze the global transcriptome of rice at the best possible resolution. Our study focused on the identification of exon-splicing junctions by relying on the deep sequencing of the transcriptome at single-base resolution by RNA-seq. We developed SPSS (searching positions of splicing sites by RNA-seq data), which we used to detect exon-intron breakpoints from paired-end short reads.
Related website
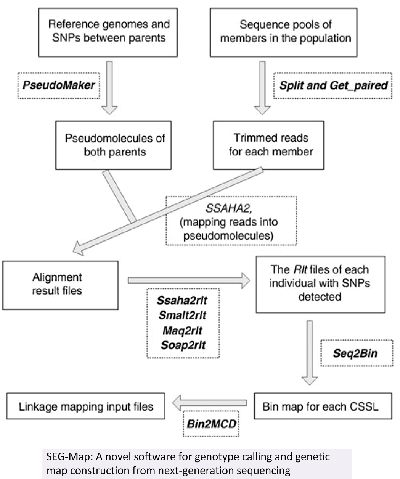
4.A sequencing-based method was recently developed to genotype recombinant populations with considerably improved resolution and reduced time and cost. To effectively implement this method, here we report the development of an analytic pipeline, sequencing enabled genotyping for mapping recombination populations (SEG-Map), for genotype calling and constructing genetic maps from next-generation sequencing data. SEG-Map was designed to interface with the commonly used tools for mapping next-generation short reads.
Related website
Selected Publications:
Researches:


1.The Oryza sativa L. indica subspecies is the most widely cultivated rice. During the last few years, we have collected over 20,000 putative full-length cDNAs and over 40,000 ESTs isolated from various cDNA libraries of two indica varieties Guangluai 4 and Minghui 63. A database of the rice indica cDNAs was therefore built to provide a comprehensive web data source for searching and retrieving the indica cDNA clones.
Related website

2.With the availability of rice and sorghum genome sequences and ongoing efforts to sequence genomes of other cereal and energy crops, the grass family (Poaceae) has become a model system for comparative genomics and for better understanding gene and genome evolution that underlies phenotypic and ecological divergence of plants. We developed a Moso bamboo genomic resource database that so far contained the sequences of 10,608 putative FL-cDNAs and nearly 38,000 expressed sequence tags (ESTs) generated in this study.
Related website

3.We applied the RNA-seq approach to analyze the global transcriptome of rice at the best possible resolution. Our study focused on the identification of exon-splicing junctions by relying on the deep sequencing of the transcriptome at single-base resolution by RNA-seq. We developed SPSS (searching positions of splicing sites by RNA-seq data), which we used to detect exon-intron breakpoints from paired-end short reads.
Related website

4.A sequencing-based method was recently developed to genotype recombinant populations with considerably improved resolution and reduced time and cost. To effectively implement this method, here we report the development of an analytic pipeline, sequencing enabled genotyping for mapping recombination populations (SEG-Map), for genotype calling and constructing genetic maps from next-generation sequencing data. SEG-Map was designed to interface with the commonly used tools for mapping next-generation short reads.
Related website

Selected Publications:
A collection of 10,096 indica rice full-length cDNAs reveals highly expressed sequence divergence between Oryza sativa indica and japonica subspecies.
Liu X, Lu T, Yu S, Li Y, Huang Y, Huang T, Zhang L, Zhu J, Zhao Q, Fan D, Mu J, Shangguan Y, Feng Q, Guan J, Ying K, Zhang Y, Lin Z, Sun Z, Qian Q, Lu Y, Han B.
Plant Mol Biol. 2007; 65: 403-15.
PMID: 17522955
Liu X, Lu T, Yu S, Li Y, Huang Y, Huang T, Zhang L, Zhu J, Zhao Q, Fan D, Mu J, Shangguan Y, Feng Q, Guan J, Ying K, Zhang Y, Lin Z, Sun Z, Qian Q, Lu Y, Han B.
Plant Mol Biol. 2007; 65: 403-15.
PMID: 17522955

Genome-wide characterization of the biggest grass, bamboo, based on 10,608 putative full-length cDNA sequences.
Peng Z, Lu T, Li L, Liu X, Gao Z, Hu T, Yang X, Feng Q, Guan J, Weng Q, Fan D, Zhu C, Lu Y, Han B, Jiang Z.
BMC Plant Biol. 2010; 10: 116.
PMID: 20565830
Peng Z, Lu T, Li L, Liu X, Gao Z, Hu T, Yang X, Feng Q, Guan J, Weng Q, Fan D, Zhu C, Lu Y, Han B, Jiang Z.
BMC Plant Biol. 2010; 10: 116.
PMID: 20565830

Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq.
Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Li W, Huang X, Han B.
Genome Res. 2009; 20: 1238-49.
PMID: 20627892
Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Li W, Huang X, Han B.
Genome Res. 2009; 20: 1238-49.
PMID: 20627892

SEG-Map: a novel software for genotype calling and genetic map construction from next-generation sequencing.
Zhao Q, Huang X, Han B.
Rice 2010; 3: 98-102.
PDF
Zhao Q, Huang X, Han B.
Rice 2010; 3: 98-102.


